flickr - ynse
DNA computing
Introduction
DNA computing is a new field of science that considering biological molecules as basic components of computing devices. It is relative to many other fields such as chemistry, computer science, molecular biology, physics and mathematics. Although its theoretical history dates back to the late 1950s, the concept of computing with molecules was only physically realized in 1994, when Leonard Adleman demonstrated in the laboratory the solution of a small instance of a well-known problem in combinatorics using standard tools of molecular biology. After this experiment, interest in DNA computing has increased dramatically, and it is now a well-established area of research.
Start of DNA Computing Technology
Lets first have a look at the starting point of DNA computing. DNA computers can’t be found at your local electronics store yet. The technology is still in development, and didn’t even exist as a concept a decade ago. In 1994, Leonard Adleman introduced the idea of using DNA to solve complex mathematical problems. Adleman, a computer scientist at the University of Southern California, came to the conclusion that DNA had computational potential. In fact DNA is very similar to a computer hard drive in how it stores permanent information about your genes.
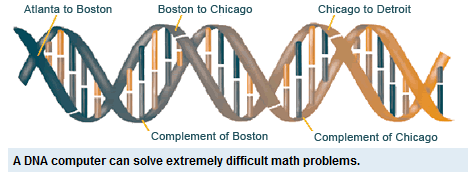
Adleman uses DNA to solve a well-known mathematical problem, called the directed Hamilton Path problem, also known as the “traveling salesman” problem. The goal of the problem is to find the shortest route between a number of cities, going through each city only once. As you add more cities to the problem, the problem becomes more difficult. Adleman used strands of DNA to represent cities. Genetic coding is represented by the letters A, T, C and G. some sequence of these four letters represented each city and possible flight path. These molecules are then mixed in a test tube, with some of these DNA strands sticking together. A chain of these strands represents a possible answer. Within a few seconds, all of the possible combinations of DNA stands, which represent answers, are created in the test tube. Adleman eliminates the wrong molecules through chemical reactions, which leaves behind only the flight paths that connect all cities. This is the moment when DNA computing is born.[1]
Though Adleman’s approach was seminal, his algorithm was not expressed within a formal model of computation. It was limited in that it was specific to solving only one problem. Also for now the present technology is stilling trying to find a way that allows biological computers to potentially change the way we do all computations.
DNA Computer Concepts
For logical consideration, it is necessary to distinguish two kinds of DNA based-computers. One is what scientists achieved today, called quasi-molecular computers (QMC) where most of the input and output operations and state transitions are driven externally by human. The other one is the final goal that scientists are desired to approach, which is called the truly molecular computers (TMC). This type of molecular computers can compute operations by self-organizing chemical reactions. So here we only discuss about QMC.
We now first take a simple example about the DNA computing system. Assume this system as a black box and we got some inputs on one side. After some kinds of convolution, outputs show up on the other side. In more details, inputs are computing problems represented by DNA sequence and they are accompanied by certain enzymes. Inside the black box are some controllable reactions which can be related to different problems. And finally after complex but fast reactions, new DNA sequences come out as outputs. Next we need to see how various problems can be fit into this black box.
As we know conventional computers deal with data in a binary form and logic gates can help us to achieve that. It is also necessary for DNA computing system but of course not using logic gates. Instead, a collection of specially selected DNA strands are used to represent various problems in binary form as DNA has a double helix structure and it is a long-term storage of information. In other words, arbitrary binary strings are encoded as strands of DNA.
Encoding
In order to make a computing system that can solve variable problems. First of all, we need to express these problems into binary forms. Actually all problems in computers are representing in binary forms because it’s easy to understand by ‘machines’. American computer scientist Richard Jay Lipton came up with an idea to encode an exponential-sized pool of starting strands, using only a polynomial number of ‘building-block’ strands. Just as Adleman encoded a large number of possible paths through a graph, using a small number of node and edge strands, a large number of assignment strands can be built using a small number of ‘variable’ strands.
An arbitrary binary string can be represented as a path through a graph, where each node represents one particular bit. Lets have a look at an example to encode a string of three bits, each represents by a node labeled A,B or C. Each node has two edges going out of it, labeled either 1 or 0. Any possible three bit string can be represented by a path going from the first node to the end. At each node the path can choose one edge, in such way the value of each bit is defined. For example, if we only take the top path at each step, we encode the string 111, and if we only take the bottom path, we end up with 000, and so on.[2] In the next step, we will focus on how the inputs are converted to outputs through the black box.
Boolean Matrix Multiplication
After the encoding process, we can now treat DNA sequence as binary strings and do some real computations. Boolean matrix multiplication which contains only ones and zeros will be a good example. Let's consider of two boolean matrixes X and Y, one is three by four and the other one is four by two. It is quite obvious that the result matrix Z after multiplication is three by two. We take the content values randomly as shown in the figure below.
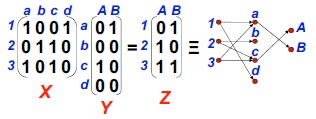
On the right hand side is a vertices and directed edges representation of the result matrix, whereas vertices represent rows and columns, directed edges represent contents in the matrix. Actually this representation also shows the process of searching and achieving the final answer. For example, first step of this computation is sum of multiplication of each element in the first row of X by each element in the first column of Y, getting result for the first element in the top row of Z and so on. As you can see, it search all the ones in first row of X and check if it matches to any ones in the first column of Y at the same position. If there is one or more matched, then it becomes one and can go further to vertex A or B respectively. If the directed edge reaches the final vertex, then result will be one and vice versa.
Now compute this example by using sets of procedures dealing with DNA strands. Express vertices as DNA strands but should be separated into two different types. Because some of them are terminal vertex however some of them are intermediate and directed edges are used to connect these two classes of vertices. Directed edge is designed to be a connector strand containing partial complementary sequences to both vertices to be connected. The graph below is more intuitive.
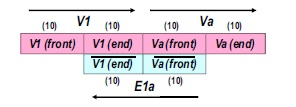
The next step is to generate the pool of all the possible combinations (paths) of vertex strands. A process called Parallel Overlap Assembly (POA) is used here. This procedure is a set of actions such as denaturing, annealing and elongation which is quite similar to PCR described in genetic engineering part. After that, filtering is required to obtain the potential strands. Here our genes of interest is those strands that start from V1, V2 or V3 and terminate at Va or Vb. Complementary terminal vertices sequences are used as primers and huge amount of result solution can be produced under PCR process. Unlike conventional method, we filter the unwanted by massively copying those we desired and reduce unwanted to a tiny proportion. Finally gel electrophoresis process is used to find out which element in the result matrix is one and then those left are all zeros.
Application - DNA Computers to Fight Diseases
Israeli scientists have developed tiny devices able to detect sighs of cancer, and release drugs to treat the disease. The work is still test-tube-based but it could lead to “nano-clinics” which remain in the body, sensing illnesses and then treating automatically. The devices are so small that roughly a trillion of them can fit into a microlitre. The research is led by Ehud Shapiro from the Weizmann Institute in Rehovot and is published in the journal Nature.[4]
This computer looks like chains consisting of three main segments. The first segment senses levels of substances which are produced by cancerous cells. It functions like a computer running through a simple algorithm. One algorithm which the team tested is intended to diagnose prostate cancer. It says that if levels of two messenger RNA molecules are lower than usual, and levels of two other are elevated, there must be prostate cancer cells in the vicinity.
If this analytical segment decides that cancer is present, it tells the second segment to release the third segment, which is an anti-cancer drug, consisting of so-called anti-sense DNA. Actually the second segment is just an operator to control releasing of the third segment. The third segment has the effect of suppressing gene activity involved in the cancer.
Prospect
Compare DNA computers to conventional computers, the former has significant advantages. First of all, the physical size is incomparable as DNA computer can be small enough to work inside a human cell. And DNA is capable of storing massive information in a small space. One cubic meter of DNA solution can be used to store billions and trillions of data with only one cubic centimeter DNA solution. Also, DNA computers will be able to perform calculations at a number of 10 to power of 20 per second, with negligible consumption of energy, only a billionth of ordinary computers today.
Remember I have mentioned truly molecular computers previously and now have a think about the cells. A cell is actually a TMC provides various functions inside human bodies. Let’s consider about the characteristics of living cells relative to conventional computers.

I believe that you all see the highly correspondences between cells and computers. That’s why DNA computing is possible and the technology in DNA field is developing so fast.
DNA computing devices could revolutionize the pharmaceutical and biomedical fields. Some scientists predict a future where our bodies are patrolled by tiny DNA computers that monitor our well-being and release the right drugs to repair damaged or unhealthy tissue. Autonomous bio-molecular computers may be able to work as ‘doctors in a cell’, operating inside living cells and sensing anomalies in the host. DNA computing research is going so fast that its potential is still emerging. This is an area of research that leaves the science fiction writers struggling to keep up.
References
[1] Martyn M.A., 2008 “DNA Computing” Invited article for the Encyclopedia of Complexity and System Science, Springer.
[2] Richard J.L., 1995 “DNA Solution of Hard Computational Problems” Science, Vol 268, Issue 5210, pp:542-545
[3] Nordiana Rajaee, Student Member, IEEE, H. Aoyagi, K. Yabe, Y. Kon and Osamu Ono, 2008 "A New Approach for Boolean Matrix Multiplication with DNA Computing"
[4] Richard R.B., 2004 "DNA Computers to Fight Diseases" BBC News Science & Environment
[5] Shungchul Ji, 1999 “The Cell As the Smallest DNA-based Molecular Computer” BioSystems 52 pp:123–133